December 22nd, 2017
ImprovementsBatch improvements
We’ve improved the batching functionality on the visual interface. In addition to the existing batching methods, you can now toggle batching on and off for any workflow on the DRAFT task page. You can also set batching criteria by any metadata, as was previously available on the API.
Learn more from our documentation.
ImprovementsData Cruncher – Suspend time
You can now set the period of inactivity after which the instance is stopped automatically and the analysis is saved.
Learn more from our documentation.
V2 Transition
On January 31, 2018, Seven Bridges will deprecate legacy projects and features on the Seven Bridges Platform. Apps, tasks, and data within your legacy projects will remain accessible. Port these to Version 2 projects to conduct further analyses.
Learn more from our post.
November 17th, 2017
ImprovementsPhenotype visualization improvement for Sonar
We’ve improved phenotype visualizations in the Sonar Playground. Use the scatterplot to visualize up to three dimensions of your query and observe trends or refine your original query. Note that this visualization is currently available only for single queries. Learn more about scatterplots for your queries.
November 3rd, 2017
Sign-on (Enterprise)
The Seven Bridges Platform now supports integration with third-party authentication services that are SAML2.0 compatible. We can quickly setup external authentication on a division level as requested by the client.
October 27th, 2017
ImprovementsAmazon KMS support for Volumes
You can now use Amazon KMS with your Amazon S3 volume. Simply provide your KMS ID when configuring your volume for the Platform. Learn more from Amazon’s documentation about using a KMS ID.
ImprovementsUpdating Help functionality
Use the Help icon on the bottom of each page to access customer support, send feedback, and find contextual supporting documentation all in the same place.
October 9th, 2017
NewSeven Bridges command line interface
We are rolling out the release of Seven Bridges Command Line Interface (version 0.5.0) which will make our public API accessible through the command line as well.
You can use the SB CLI to leverage the functionalities of our API to interact with the Platform. You can also automate your interaction with the Platform by using the SB CLI in scripts or programs.
SB CLI is available for Linux, macOS, Windows, and FreeBSD.
Learn more from our documentation. Or, consult the CLI reference.
ImprovementsOne second billing
We’ve switched billing for AWS EC2/EBS as well as Google instances to 1 second increments. Change will also take effect on EU platform. There is also 1 minute minimum charge per-instance. Prices in pricing plans will remain the same (per hour) but will be billed per second. Learn more from our documentation.
September 4th, 2017
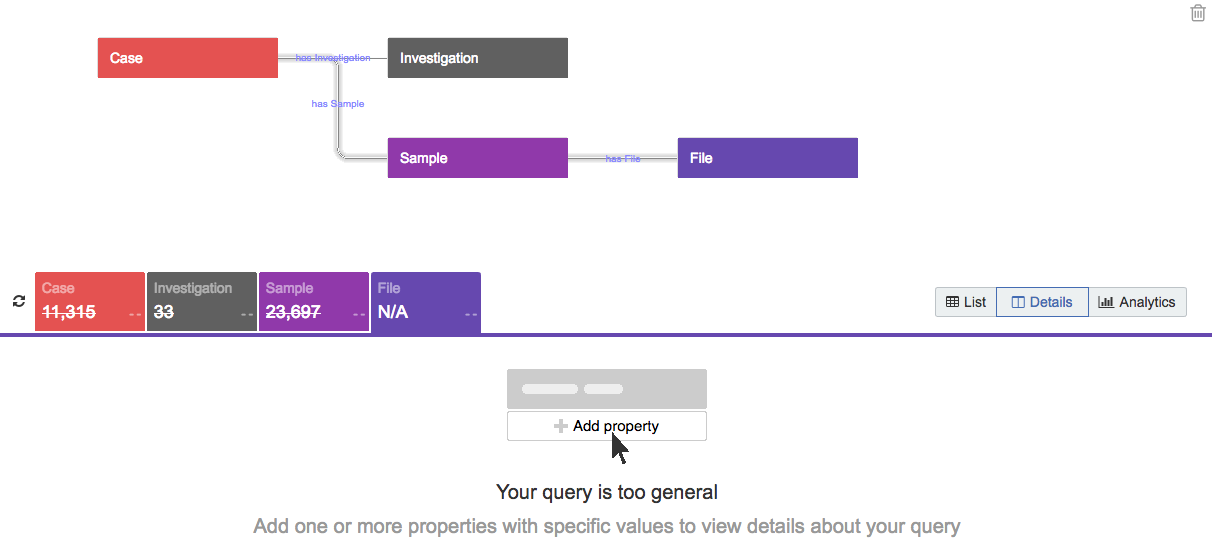
NewQuery validation
We’ve implemented a system to help you generate queries with a meaningful sample size of data. For instance, if you add more than three entities (e.g. Case, Sample, File) without applying any filters, the Data Browser prompts you to add one or more properties with specific values to narrow your query before continuing, as shown below.
The Data Browser similarly prompts you to add filters if you create a query that has symmetrical branches without any filters applied, as shown below.
Learn more about adding filters to your query.
ImprovementsTable of results improvements
The table below the Data Browser contains further details about your query. In addition to the List view and Analytics view, we’ve introduced a Details view.
The Details view displays details about a selected entity in the context of your query and consists of three panels. The first panel details the inbound and outbound connections for the selected entity in the context of your query. The second panel displays a list of UUIDs corresponding to that entity that match your query. If a specific UUID within the second panel is selected, the third panel displays the metadata corresponding to it.
This new Details view enables you to explore the connections and relations between entities in your query more easily and thus ensure that your query is identifying the data you need to meet your research goals.
Learn more about the Details view.
August 28th, 2017
ImprovementsSupport for Amazon Web Services Spot instances
Seven Bridges has introduced support for Spot instances on the Amazon Web Services (AWS) deploy of the Platform. Spot instance support can be selected as a default for projects and and an option for each task execution. By selecting a spot instance execution costs can be dramatically reduced. Our testing indicates an execution cost savings of over 75% on common workflows.
Due to the nature of how AWS handles Spot instances, they can be interrupted while tasks are running. If a Spot instance is interrupted, Seven Bridges’ job retry functionality will automatically restart interrupted and remaining unfinished jobs on an On-Demand instance to prevent further interruptions. Such an interruption may impact the cost savings from using a Spot instance and can result in a longer overall runtime, but the reliability of task execution is unaffected.
For more information see blog post and the feature documentation.
NewSBFS [Beta release]
SBFS is a command line tool which enables interaction with Platform project files that are mounted as a local file system.
Use SBFS to make project files available on a local file system and thus as accessible as any other locally available file. This eliminates the need for downloading complete files to a local machine, which is especially useful when working with large files exceeding the size of a local disk. With SBFS, parts of a file are accessible without necessitating a complete file download and users can perform interactive analyses on a local machine (or server instance) without needing to bring their tool to the Platform.
SBFS is available for Linux and macOS operating systems, and beta version is available for download from the new Data tools page.
Learn more from SBFS documentation.
ImprovementsDatasets improvements
TARGET GRCh38 Dataset
The Therapeutically Applicable Research to Generate Effective Treatments (TARGET) dataset provides genomic, transcriptomic, and epigenomic data from patients representing several childhood cancers and serves as a valuable complement to the existing genomic and multi-omic datasets available on the Seven Bridges Platform via the CGC. The complete TARGET GRCh38 dataset, which includes both Open Data accessible to all researchers and Controlled Data, to which access is regulated by the Database for Genotypes and Phenotypes (dbGaP), is now available on the Platform. This dataset can be queried using the Data Browser to generate custom cohorts from within this dataset as well as cohorts derived from multiple similarly aligned datasets such as the TARGET GRCh38 and TCGA GRCh38 datasets.
TARGET GRCh38 dataset and its metadata.
CPTAC Dataset
The Platform now provides access to mass spectrometry data that were generated by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) as part of the TCGA initiative to characterize and quantify the proteome of cancer samples. This dataset represents 335 samples from patients with Breast Invasive Carcinoma, Colon Adenocarcinoma, Ovarian Serous Cystadenocarcinoma, and Rectum Adenocarcinoma for whom matched genomic data are available. The dataset can be queried using the Data Browser to generate custom cohorts from within this dataset as well as multi-omic cohorts across the TCGA GRCh38 genomic and CPTAC proteomic datasets.
Learn more about the CPTAC data, and CPTAC metadata.
Multiple-dataset Querying
To maximize the accessibility and value of the multi-omic datasets available on the Platform, the Data Browser now enables cross-dataset queries for datasets with harmonized metadata. This allows researchers to use the Data Browser to identify cohorts of interest across multiple genomic datasets such as the GRCh38 alignments of TCGA and TARGET.
Learn more about cross-dataset queries through a sample query.
August 1st, 2017
NewElastic Block Storage customization feature
Overview
On August 1, we released a feature that provides you with the ability to customize the amount of Elastic Block Storage (EBS) disk space attached to different Amazon instance configurations. EBS customization is useful for bioinformatics workflows because it provides the ability to optimize your computation by giving you greater control over requirements and costs.
To learn more about EBS customization, please see our documentation.
Passing through EBS charges from AWS
Along with the introduction of EBS customization, there will be changes in the way that you are billed for Elastic Block Storage and Amazon Web Services (AWS) costs on the Platform. Starting on August 1, you will see an additional charge on the task overview page and your bill when you use EBS disk space. Information about the instance and EBS usage cost is available on the tooltip on the task overview page, next to the total price. Before August 1, you will see a $0 charge for attached disk space.
Why will I be charged more when I use EBS disk space?
When AWS introduced EBS they changed their pricing structure to charge separately for the compute and storage. AWS charges ~$0.10 per GB*h/month for EBS disk space. Up to now, Seven Bridges has paid costs for EBS usage on your behalf, however starting on August 1, along with the capability to fully customize EBS, we will begin passing through EBS costs. Our policy is to to be completely transparent around your AWS charges and to not charge a premium to access AWS services through the Platform.
Check out our documentation for more information on EBS charging.
April 14th, 2017
NewSimons Genome Diversity Project (SGDP) Dataset
The Simons Genome Diversity Project (SGDP) Open Access dataset contains complete genome sequences from 130 diverse human populations. It is the largest dataset of diverse, high-quality human genome sequences ever reported and includes many deeply divergent human populations that are not well-represented in other datasets, which makes the SGDP dataset ideal for interrogating the genomic landscape of different populations.
This dataset is available for analysis under the Public projects tab of the top navigation bar. You won’t pay for storage of the raw data files: copy the entire project or select files into your own project on the Platform to take conduct further analysis. Or, compare SGDP data alongside of your own data.
Learn more about using the SGDP public project.