February 20th, 2026
Enhanced Data Studio Session Backup and Shutdown
General
This release includes stability enhancements to Data Studio session shutdown and backup, reducing the occurrence of stuck sessions and improving the reliability of analysis recovery.
Improvements
The reliability of the process that runs when you stop a Data Studio session has been improved. The updated logic better handles large and complex analysis environments, ensuring that backup and cleanup steps complete more consistently before a session is terminated.
Fixed Bugs
Several issues that could cause Data Studio sessions to remain in a saving or stopping state for extended periods and result in incomplete backups have been resolved. The following underlying problems were fixed:
- Locked files during backup: Improved handling of files that remain in use while the backup is running, so they no longer prevent proper cleanup and completion of the backup process.
- File upload reliability: Strengthened error handling for timeouts when uploading large files, reducing incomplete transfers that could interrupt session backup.
- Complex environment handling: Fixed failures when saving sessions with many installed packages and dependencies, improving stability for complex analysis environments.
- System metrics handling: Fixed an issue where unexpected internal data could interrupt the backup workflow.
- Bulk file operations: Optimized how large batches of files are processed during backup, preventing platform slowdowns during session shutdown.
Resources
If you experience issues with stopping or recovering a Data Studio session, please contact your Velsera Seven Bridges representative or email support@velsera.com.
Learn more about Data Studio.
December 18th, 2025
Recently Published Apps
- CellTypist: Automated cell type annotation in single-cell RNA-seq data. It uses pre-trained machine learning models on curated datasets to deliver fast, standardized, and reproducible cell labeling. This is especially valuable for large-scale immunology and tissue profiling studies, reducing manual effort and subjectivity.
- GATK VariantToTable: Transform raw VCF data into clean, structured tables. Select the INFO, FORMAT, or genotype fields you need, and the tool generates a ready-to-use CSV/TSV file for effortless filtering, visualization, and downstream analysis in Excel, R, and Python.
- GATK GenomicsDBImport: Efficiently combine multiple gVCF files into a GenomicsDB workspace, enabling scalable joint genotyping. It organizes variant data in a tiled database format, which allows faster queries and reduces storage requirements compared to raw gVCFs. This step is typically performed before running GenotypeGVCFs in large cohort analyses. The updated version 4.6.2.0, now featuring a DB update option.
- The MCmicro workflow: A scalable and reproducible pipeline for processing multiplexed tissue imaging data. It integrates tools for image registration, illumination correction, segmentation, and feature extraction. These tools produce high-quality single-cell spatial datasets and are designed for large-scale studies, MCmicro standardizes analysis across experiments, enabling downstream biological insights from multiplex imaging platforms. This updated workflow version, with existing models and support for custom models.
November 4th, 2025
Enable Project Download Restrictions Retroactively
This release introduces the ability to apply download restrictions to a project retroactively. Previously, the download restriction could only be applied while creating a project. With this release, the admins and project creator can modify the project settings to enable download restrictions at anytime.
The Download Restriction feature has been upgraded to also be applicable retroactively to existing projects, without recreating projects or migrating files. This enables enhanced restriction of data access on the platform, aligning with security policies.
- The restriction is irreversible, ensuring consistent security policies.
- Internal teams can support rollback based on requests, ensuring flexibility without compromising control and reducing time
November 4th, 2025
Enabling new AWS families with GPU-enabled Instances on Data Studio
This release introduces adding new AWS families with GPU enabled Instances within Seven Bridges Data Studio. It enables users to perform advanced medical image visualization, segmentation, GPU acceleration and secure data integration.
Enabled AWS Instances on Data Studio
We are thrilled to announce that we have enabled new AWS families with GPU enabled Instances on Data Studio to provide even more options for optimizing your computational workloads.
New AWS Instance Families Include:
- Compute-Optimized: C7i, C6i
- Memory-Optimized: R7i, R7iz, R6i, R6in, X2idn, X2iedn, X2iezn
- General Purpose: M7i, M6i
- GPU-Enabled: G6, G5
This update empowers researchers and analysts to execute high-performance computational tasks directly from Data Studio without external infrastructure setup.
It simplifies workflow execution, reducing processing time and improving efficiency for GPU-dependent applications.
November 4th, 2025
3D Slicer now available in Data Studio
General
This release introduces 3D Slicer as a fully integrated analysis environment within Data Studio, enabling advanced medical image visualization and segmentation.
New feature
3D Slicer can now be launched directly from Data Studio for interactive medical imaging and 3D visualization workflows, with the option of GPU-powered compute instances for high-performance rendering. This release enables you to perform entire imaging workflows from data import, visualization, annotation within the secure and compliant Seven Bridges platform, and eliminates the need to move sensitive data across multiple tools or local machines.
October 30th, 2024
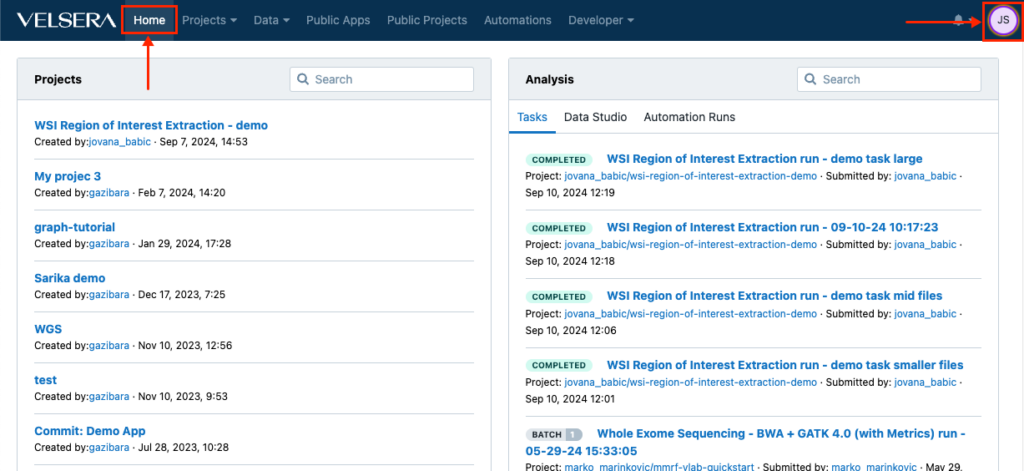
Changes in the main navigation
The main navigation now features a Home button which returns you to the dashboard at any given point. In addition, the icon for accessing the account settings now shows your initials instead of the username.
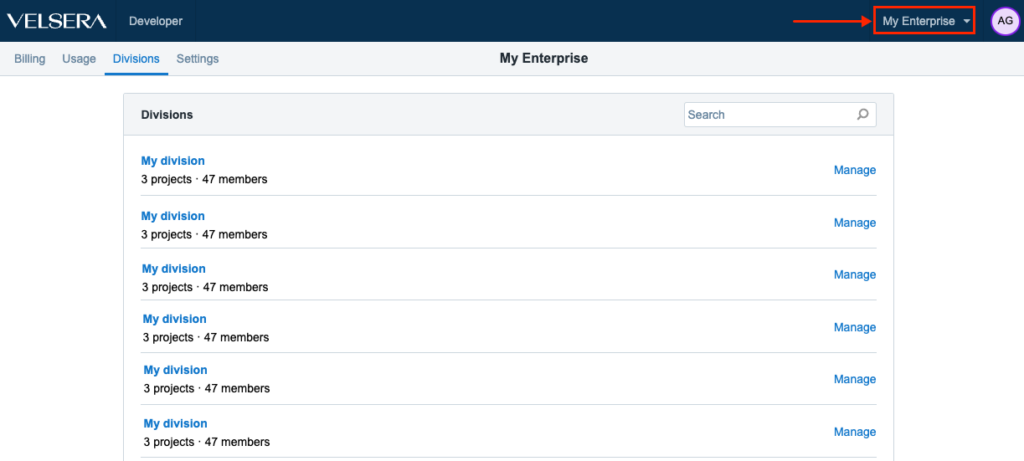
Improved UX for Enterprise users
We have introduced a small change in order to improve and optimize the UX for the Enterprise users. The menu for accessing Divisions is now in the upper right hand side, right next to the Account Settings.
October 28th, 2024
Nextflow – enhanced execution and scalability
We’re excited to announce powerful new features to enhance Nextflow performance, scalability, and resource optimization. These updates streamline workflow execution and expand support for advanced configurations, making it easier than ever to manage complex pipelines.
Multi-Instance Execution
Support for multi-instance execution allows Nextflow workflows to leverage multiple instances concurrently. This improvement enables increased parallelism and better resource utilization across workflows, resulting in significant performance boosts for complex or large-scale data pipelines.
Memoization (Work Reuse)
Introducing memoization for workflow tasks, allowing Nextflow to reuse previous results, significantly reducing compute time and resource usage. This feature is especially beneficial for iterating on workflows with minor changes, as previously executed tasks are reused when input data and parameters remain consistent.
Elastic Disk Support
Elastic disk capability allows dynamic scaling of disk resources based on workflow requirements. This flexible storage management ensures that high-demand workflows have access to the necessary storage without manual intervention, optimizing both performance and cost.
Instance Hints for Optimized Resource Selection
Nextflow now supports instance hints to provide tailored recommendations for instance types based on workflow needs. These hints help reduce costs and improve runtime efficiency by selecting the most suitable instance types for different tasks.
Real-Time Job Monitoring for Worker Instances
Enhanced job monitoring capabilities provide real-time insights into the status of worker instances. Users can monitor task progress and troubleshoot issues as they arise, leading to faster adjustments and a more seamless workflow execution experience.
Compatibility with All Nextflow Executor Versions (Post 21.10.0)
Full support for all Nextflow executor versions after 21.10.0 allows users to run workflows with the exact executor version they need, ensuring compatibility and reproducibility across executions. This enhancement allows teams to replicate their work consistently while taking advantage of continuous improvements in Nextflow ecosystem.
June 10th, 2024
Recently published apps
Somatic small variant callers for long read data, ClairS (0.2.0) and ClairS-TO (0.1.0) (for matched tumor-normal pairs and tumor-only data, respectively) have been published to the Seven Bridges Platform.
May 13th, 2024
Recently published apps
We have published the GCTA 1.94.1 tool on the Seven Bridges Platform. GCTA is a suite of tools for various genetic analyses using genome-wide data. GCTA (Genome-wide Complex Trait Analysis) was initially developed to estimate the proportion of phenotypic variance explained by all genome-wide SNPs for a complex trait but has been greatly extended for many other analyses of data from genome-wide association studies (GWASs)