3-hour Whole Genome Analysis with GATK4
In the pursuit of accelerating next generation sequencing data processing for clinical applications, Seven Bridges has developed a configurable GATK4 workflow 3.0 times faster than previous iterations. Following up on our initial push of the GATK4 workflow in 2017, and our recent update with the Broad’s Best Practices, we’ve worked to reduce workflow runtime on the Seven Bridges Platform by introducing configurable parallel instances.
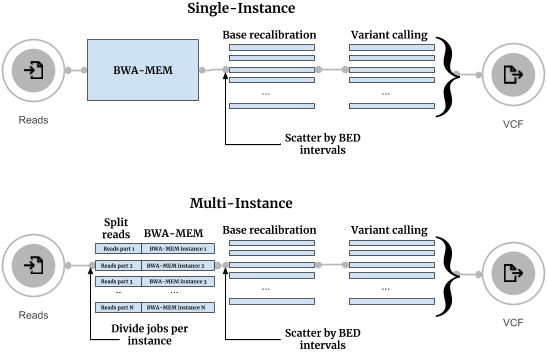
Conceptually, in the new workflow, which also follows Broad’s Best Practices, sequence reads are split into an optimal number of groups, and each group is processed independently on an individual instance (Figure 1). After processing, all results are merged together. Variant call quality is equivalent between our Multi-Instance and Single-Instance GATK 4.0 Whole Genome workflows.
The execution flows of the Single-Instance and Multi-Instance GATK 4.0 Whole Genome Sequencing workflows are shown below:
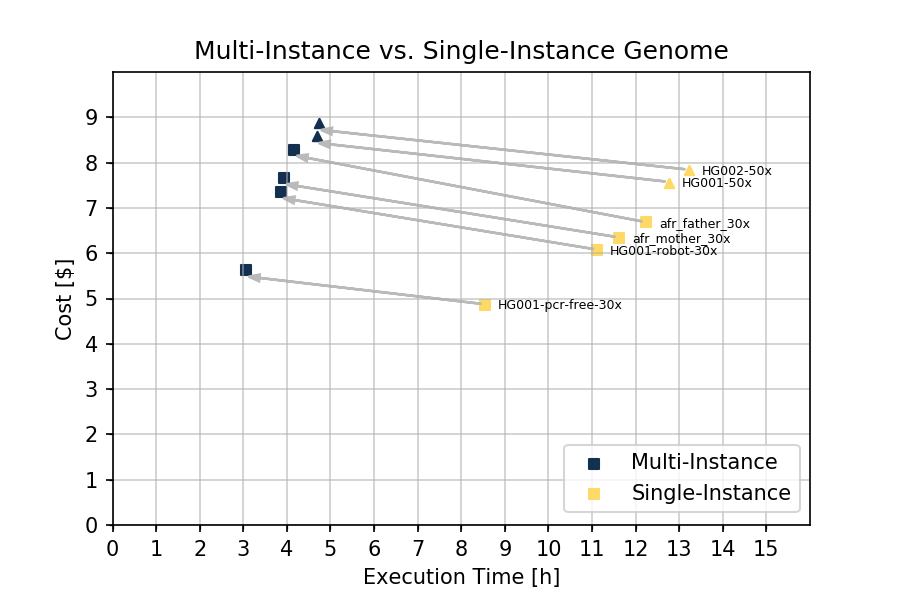
The improvements in execution time are shown in the figure below:
The execution time for a 30x whole genome through the Single-instance Workflow (n=4) varies between 8.5 and 13 hours (11.28 hours on average) with 1000 Genome and Genome In A Bottle samples (Figure 2). The Multi-Instance Workflow varies between 3 and 4 hours (3.75 hours average) achieving a 3.0x speed improvement over previous version on the Seven Bridges Platform. With the Multi-Instance Workflow the average cost increased from $6.20 to $7.20 on amazon spot c4.8xlarge instance (36 CPUs, 60 GB). For 50x samples (n=2), we observed a 2.8x speed improvement, from 13 hours to 4.7 hours, with average cost increasing from $7.69 to $8.72
In order to validate variant call quality between the Single-Instance and Multi-Instance GATK 4.0 Workflows we compared results using Genome In A Bottle (HG001 and HG002) samples. The differences in precision, recall and f-score were smaller than 0.001%, which is expected due stochastic effects (data not shown).
The final stage of all previous GATK workflows deployed on the Seven Bridges Platform was Variant Quality Score Recalibration. With GATK 4.0, it is recommended to exclude VQSR for single sample processing. For that reason, we developed a Smart Variant Filtering tool for filtering single whole genome samples. This tool has been added to our Multi-Instance GATK4 workflow to increase the precision and f-score of variant calls
In summary, this new BWA/GATK4 workflow on the Seven Bridges Platform offers a configurable way for users to employ parallelization to achieve reduced run times for time-sensitive applications without losing accuracy or incurring substantial additional costs.