Go beyond somatic variant calling: GATK Somatic SNVs and INDELs (Mutect2) 4.1.9.0
The GATK Somatic SNVs and INDELs (Mutect2) 4.1.9.0 workflow is a somatic variant caller workflow that uses local assembly and realignment to detect single nucleotide variants (SNVs) and insertion and deletion (INDEL) changes. This Mutect2 tool (see the original publication on BioRxiv) is an improvement upon the original “MuTect” tool created by Cibulskis et al, detailed in the Nature Biotechnology article publication here in 2013. Mutect2 is an improvement upon the original MuTect by introducing the use of the HaplotypeCaller assembler.
The Mutect2 tool itself, together with a few other tools, are compiled into this Mutect2 workflow (GATK Somatic SNVs and INDELs 4.1.9.0). This workflow is capable of operating in multiple modes: it can be used to detect SNVs and INDELs in one or more tumor samples from a single individual, with or without a matched normal sample. Here, assembly implies whole haplotypes and read pairs, rather than single bases, as the individual units of biological variation and sequencing evidence, with the effect of improving variant calling. Beyond local assembly and alignment, the Mutect2 tool is based on several probabilistic models for genotyping and filtering that work well for all sequencing depths.
Using this workflow for your research
The Mutect2 workflow runs on a single tumor-normal pair or on a single tumor sample and performs additional filtering and functional annotation tasks. The typical use-case for the Mutect2 workflow is to detect somatic variants present in a tumor sample. The Mutect2 workflow can also be used for mitochondrial variant calling, and detection of somatic mosaicism.
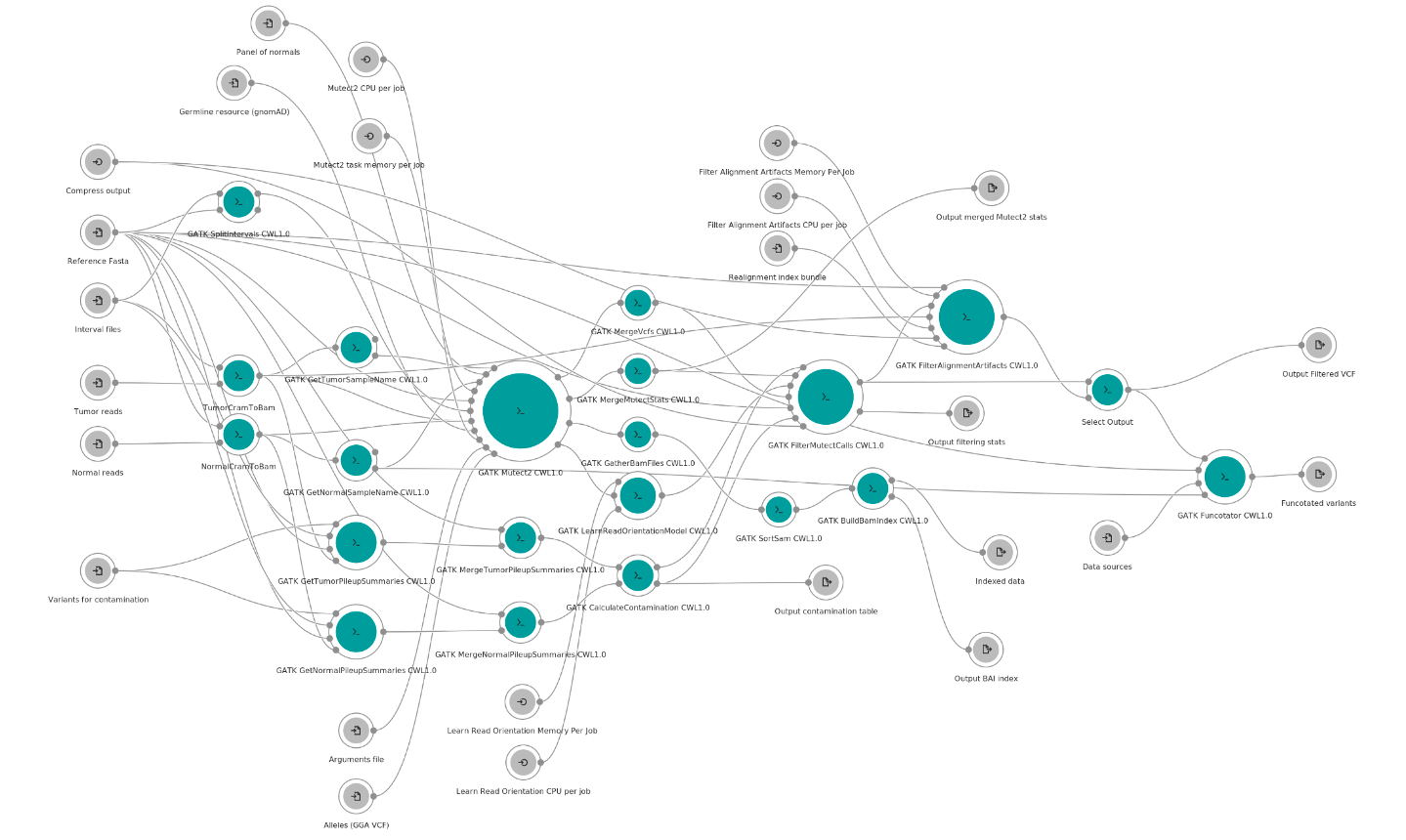
Figure 1: Overview of the GATK Somatic SNVs and INDELs (Mutect2) 4.1.9.0 workflow available on the Seven Bridges platforms.
For more details on the specific inputs and outputs for this workflow, and for a zoomable version of this figure, please see its description page on NHLBI BioData Catalyst and on the CGC.
Implementation in the Common Workflow Language v1.0
The GATK team from the Broad Institute originally created this workflow and made it publicly available in the Workflow Description Language (WDL) format, see here. WDL is a convenient way to represent data processing workflows in a human-readable way. Seven Bridges instead uses the Common Workflow Language (CWL), a widely supported, open-source specification for workflow descriptions. The GATK’s WDL description of this Mutect2 workflow was used to create our implementation in CWL v1.0 format, without making any changes that will constitute a significant departure from the WDL version. This was confirmed by obtaining the same results for the workflow’s performance benchmarking for both WDL and CWL versions. There is a wide variety of executors that support CWL, and workflows created in CWL are highly portable and reproducible. A given CWL app or workflow is not restricted to the Seven Bridges environment: they can be run on a laptop, high-performance computing clusters, and on raw cloud infrastructure, among others.
Execution Time and Cost
At Seven Bridges, our Bioinformatics Team performs benchmarking on many of our tools and workflows, in order to give users a better understanding on execution time and cost. In the table below, we represent the runtime execution cost in comparison to the unmapped BAM file size. The runtime of this workflow on the Seven Bridges cloud infrastructure varies proportionally with the size of the input files. The size of an unmapped BAM file containing raw sequencing reads has the largest influence on workflow execution time.
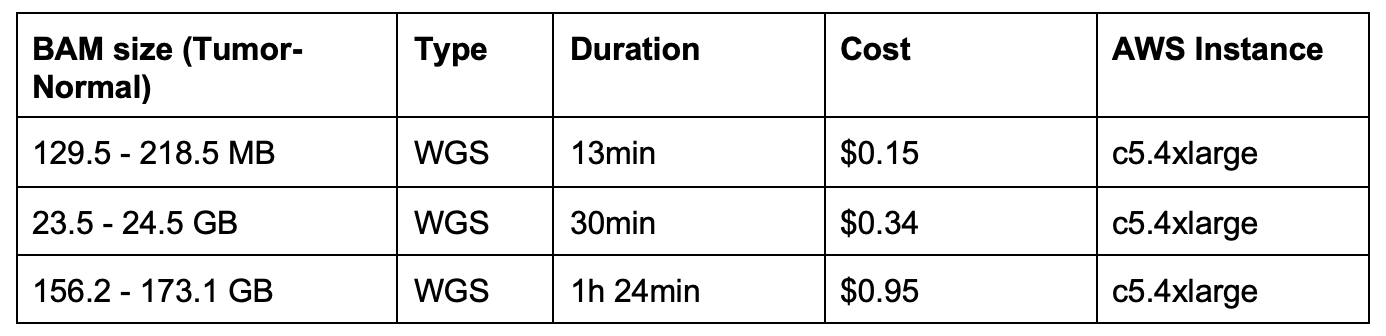
The price of execution for this workflow varies proportionally to input sample size, which could be significantly reduced (up to 75%) when using Amazon Web Services (AWS) spot instances. To learn more about spot instances, visit our Knowledge Center.
Running on the Seven Bridges platforms
We encourage you to explore somatic variants in cancer data using our GATK Somatic SNVs and INDELs 4.1.9.0 workflow. A good starting point could be investigating variants in one of the many popular datasets hosted on the Seven Bridges platforms, such as Therapeutically Applicable Research To Generate Effective Treatments (TARGET) on the Cancer Genomics Cloud and the Trans-Omics for Precision Medicine (TOPMed) program on NHBI BioDataCatalyst powered by Seven Bridges. In addition to our hosted datasets, we also feature a variety of methods to easily upload your own data to the Seven Bridges environment, and to run the subsequent analyses using our cloud infrastructure. For more information on how to get started on the Seven Bridges platforms, contact us today.