The GA4GH Workflow Execution Challenge: evaluating reproducibility and portability across platforms
Deploying portable analysis for large genomics projects
Large-scale, geographically distributed genomics analysis efforts, such as the PanCancer Analysis of Whole Genomes (PCAWG) project, highlight a trend in our community towards moving compute to where data resides. For PCAWG, this was out of necessity as the genomes for ~2,800 cancer donors were stored across 8 different locations around the world. There are many reasons for moving compute to data, ranging from the expense of hosting multiple copies of large datasets, to the time needed to transfer across the public Internet, as well as legal limitations on data mobility. To enable large-scale scientific discovery, we need to be able to use workflows that are portable, reproducible, and can run on the desired data regardless of the physical location.
Recent advances from the field of Information Technology (IT), such as cloud infrastructure, software containers, and workflow description languages, have allowed projects such as PCAWG to get closer to a realization of the vision of seamless mobility of algorithms. Cloud technologies, with commercial offerings from Amazon, Google, Microsoft and others, along with private cloud solutions such as OpenStack, allow researchers to dynamically size computational and storage infrastructure programmatically. Platforms like Seven Bridges makes the process accessible for researchers with less background in IT.
Complementing cloud infrastructure is the use of software containers, the most popular being Docker with related projects such as rkt and Singularity. These components allow researchers to create lightweight virtual environments into which they can install software, configuration, and small data files. The resulting images can easily and quickly be shared through a variety of platforms and present a fantastic way to reliably move tools across different computational environments with little effort or danger of incompatibility. Building on the functionality and success of Docker and other containers, projects like the Common Workflow Language (CWL) and the Workflow Description Language (WDL) enable scientists to express complex workflows that string multiple tools together, provide clear definitions of inputs and outputs, runtime requirements, and enable patterns commonly seen in genomics analysis such as scatter and gather. What makes CWL and WDL incredibly useful is their decoupling from execution environments. Workflows written in one of these languages can work on an increasing number of commercial and open source platforms.
Developing GA4GH API standards for cloud workflows
The work of the Global Alliance for Genomics and Health (GA4GH) complements the technologies of clouds, containers, and standardized workflow descriptors through the creation of Application Programming Interface (API) standards. In particular, the Cloud Work Stream (formerly the Containers and Workflow Task Team), is developing a series of APIs for standardizing how containerized tools and CWL/WDL workflows are exchanged; how tasks are executed across clouds; how workflows are executed in various platforms; and how inputs and outputs are managed in a cloud-agnostic fashion. These emerging standards are based on the needs and experiences of projects like PCAWG, a series of large driver projects that have volunteered to participate in the standards creation process, and interested community members.
The standards being developed by GA4GH are key to enabling frictionless compute on data distributed on clouds, both private and public, located around the world. This ability is critical to enabling scientific discovery across growing global datasets and collaborations. However, despite the increasing commoditization of compute, excellent containerization technologies, and emerging API standards for linking them together, the real proof that we have achieved this vision rests on clear demonstrations. To facilitate this, GA4GH partnered with the DREAM Challenges and Sage Bionetworks to create a series of challenges that test our community’s abilities to make and use portable tools and workflows.
The Tool Execution Challenge
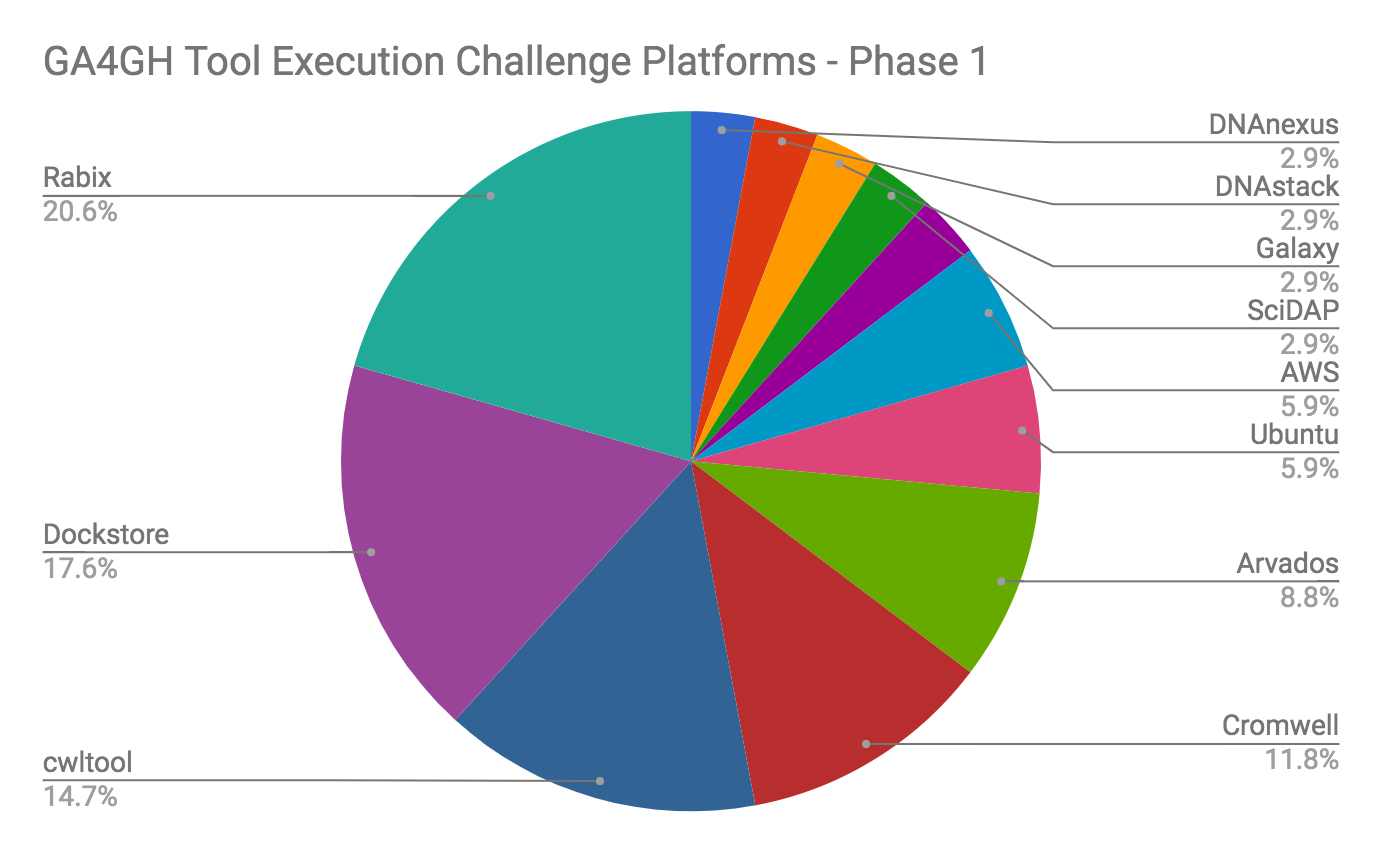
Phase I of the challenge (https://www.synapse.org/GA4GHToolExecutionChallenge) started with creating a very simple, Docker-based tool (md5sum) and posting it to the Dockstore, a repository that redistributes tools and workflows using the GA4GH Tool Registry Service standard. In this challenge, participants retrieved the tool from Dockstore (including both the Docker image and the CWL/WDL descriptor that described how to call the tool) and executed it in the platform of their choice. A total of 35 participants were successful in this activity, producing the correct md5sum for the test datafile. The platforms used ranged from community solutions like cwltool and Cromwell to commercial platforms. Regardless of its simplicity, the challenge demonstrated in a tangible way that a containerized tool described with CWL/WDL could be successfully passed from researcher to researcher and used without modification on a variety of execution platforms, producing the expected result.
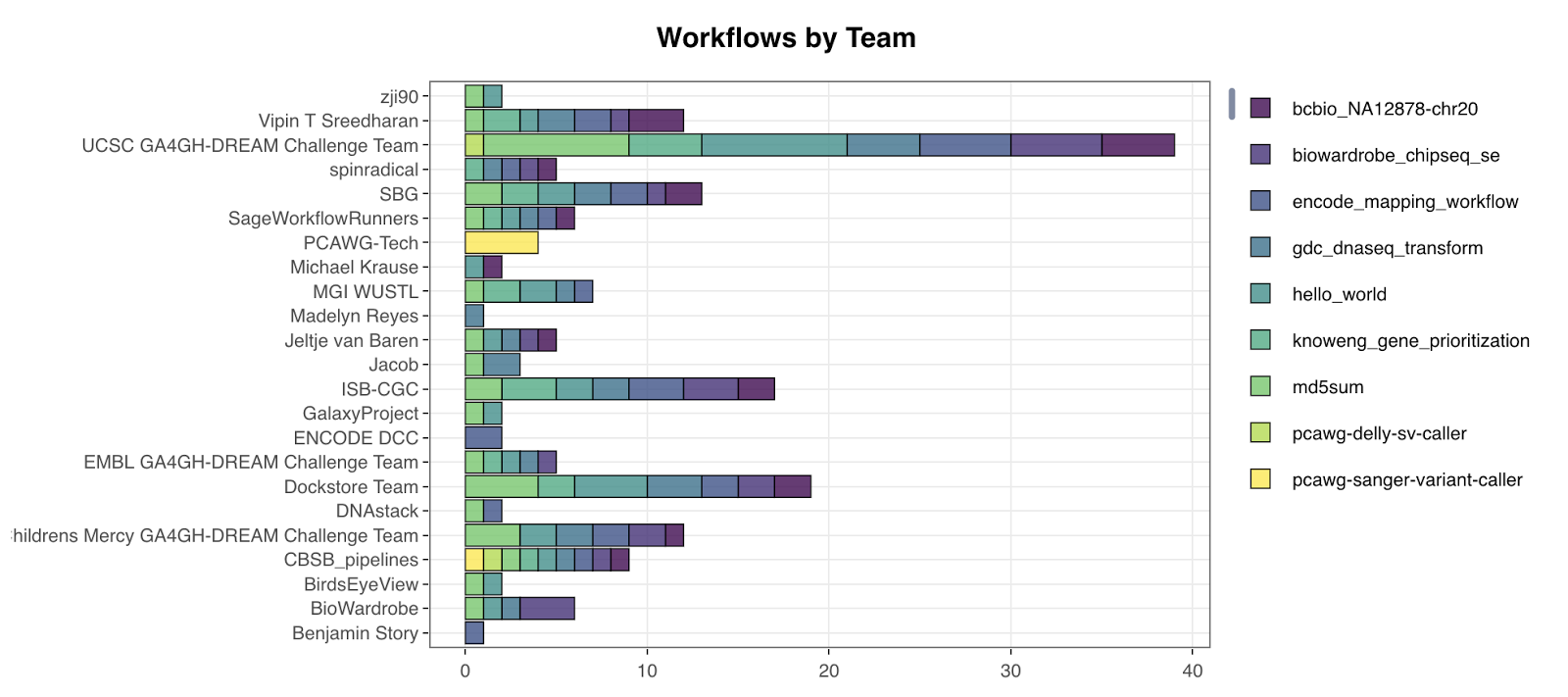
Phase II of the challenge (https://www.synapse.org/WorkflowChallenge) is ongoing and set to wrap by the end of 2017. It picks up where Phase I ended and attempts to show reproducibility/portability across platforms by using complex, real-world workflows that researchers already use in large-scale projects. Unlike a simple md5sum tool, these workflows represent best practices from the Genomic Data Commons, Broad Institute, ENCODE, PCAWG, and other large projects. In order to successfully participate in the challenge, one or more workflows are executed on the participant’s platform of choice using test data provided by the workflow author. The resulting output is then checked by a tool provided by the workflow author, which is capable of judging output concordance with the “known good” for that workflow test dataset. Participants that successfully pass the concordance check then upload their output to Synapse and document how they ran the workflow in the platform of their choice. Results are still being collected but currently the challenge has 112 registered participants and has received 176 successful submissions for 9 workflows.
Looking forwards
Ensuring that the standardized workflow descriptions for phase II of the challenge run seamlessly across execution environments, required collaboration between workflow authors and the platforms where these workflows are executed. For example, Seven Bridges worked closely with workflow authors to achieve this on the Cancer Genomics Cloud and with Rabix, theirour open source toolkit for creating and executing CWL. The GA4GH-DREAM Workflow Execution Challenge would love to have as many Seven Bridges users as possible participate, as part of our goal is test these workflow descriptions on as many platforms as possible, with multiple submissions from each platform. You can learn how to register and participate in the challenge here.
Once you’ve registered you can click on the links below to find instructions on how to execute each of the workflows on Seven Bridges Cancer Genomics Cloud or using Rabix. Help us demonstrate workflow portability and learn how to use some of the cutting edge workflows provided by the community.
With the success of Phase I, and the growing participation in the ongoing Phase II, we think there is a recognition by the community of the importance to directly test portability across systems. We are currently planning a series of additional challenges that will directly assess the ability to run analysis programmatically over GA4GH APIs for task execution (Task Execution Service, TES), run whole workflows on remote systems (Workflow Execution Service, WES), and read/write data objects across cloud storage systems (Data Object Service, DOS). In addition to future phases for the GA4GH-DREAM Challenges, we view this activity as being highly related and relevant to other community efforts to demonstrate tool portability and interoperability including the NIH Data Commons efforts. We hope that we can combine efforts into large, inclusive challenges that span a wide variety of projects and platforms.
Seven Bridges platform executions:
BCBio Variant calling – synapse wiki
Biowardrobe Chip-Seq – Not available yet
GDC DNA-Seq – synapse wiki
Hello world – synapse wiki
MD5 sum – synapse wiki
ENCODE Chip-Seq – synapse wiki
KNOWENG Gene prioritization – synapse wiki
Rabix executions:
BCBio Variant calling – synapse wiki
Biowardrobe Chip-Seq – synapse wiki
GDC DNA-Seq – synapse wiki
Hello world – synapse wiki
MD5 sum – synapse wiki
ENCODE Chip-Seq – synapse wiki
KNOWENG Gene prioritization – synapse wiki