Manifest-based DRS import: A practical solution for cross-dataset analysis to empower translational research
A key challenge in data discovery is the coordination and assembly of datasets spanning multiple from across Data Coordination Centers on a cloud compute platform in a way that is both easy to use and meaningful.
To meet this challenge, we have implemented a GA4GH Data Repository Service (DRS) manifest-based import for Common Fund Data Ecosystem (CFDE) Data Coordination Centers (DCC) on our CAVATICA platform. This DRS manifest-based import connects the associated metadata and provides a harmonized computational framework that allows a user to create a cross-Common Fund dataset cohort which, in turn, has the potential to accelerate platform-based discovery and clinical translation.
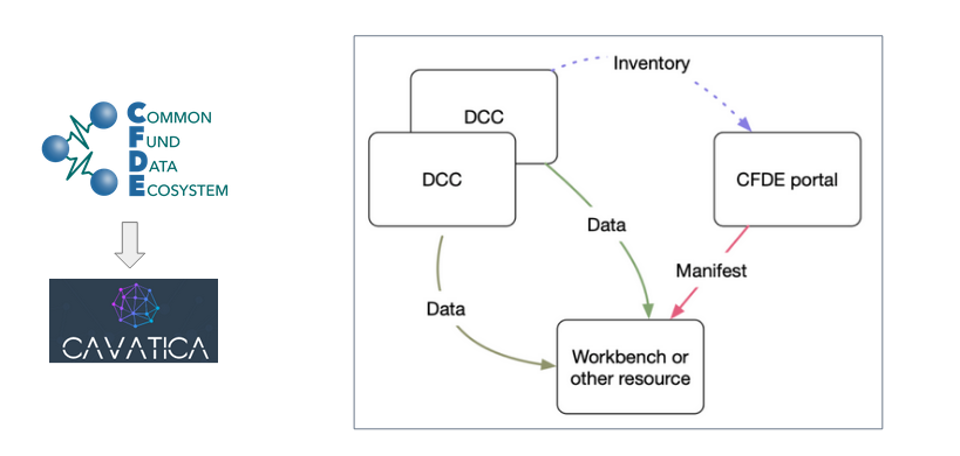
The combination of interoperable metadata and harmonized computational framework sets the stage for cross-DCC analyses and tool development, including for gene expression analysis. To import data into CAVATICA using the DRS standard, a researcher can create a manifest from the CFDE portal with harmonized GTEx and Kids First neuroblastoma RNA sequencing assays then import it into a collaborative CAVATICA workspace. The manifest is created using NCPI file manifest standards. The user’s dbGaP access authorizations are checked by CAVATICA and the data becomes accessible only if the user has proper authorization.
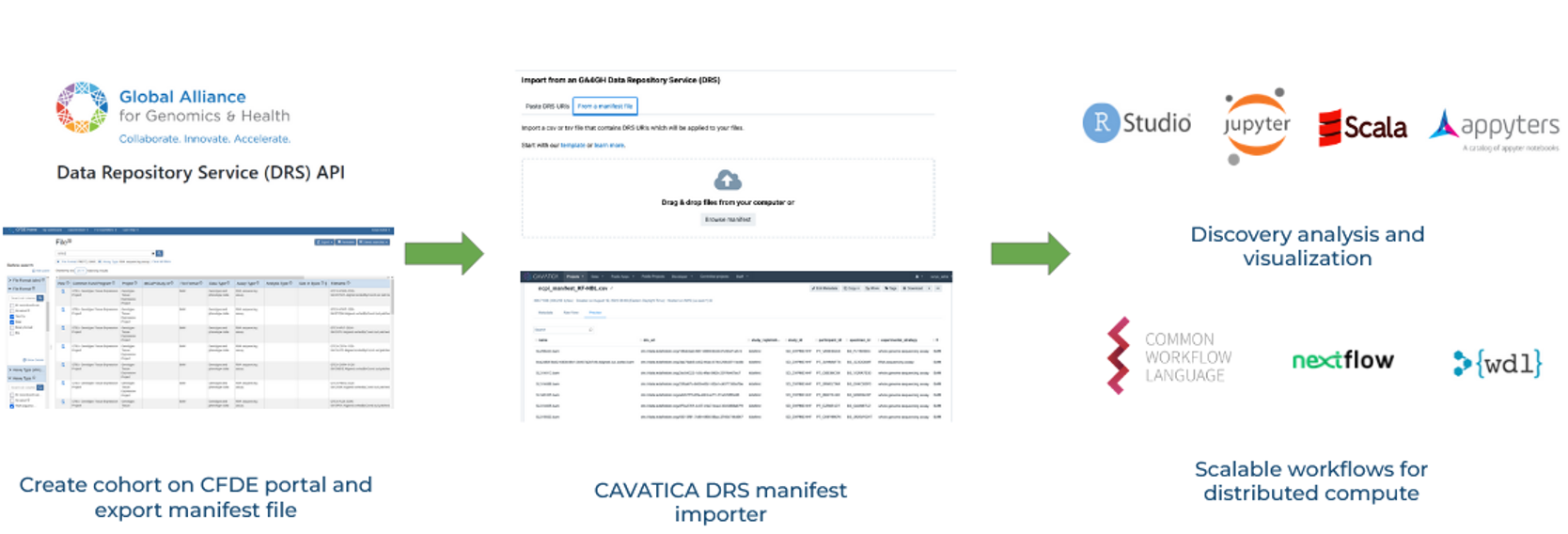
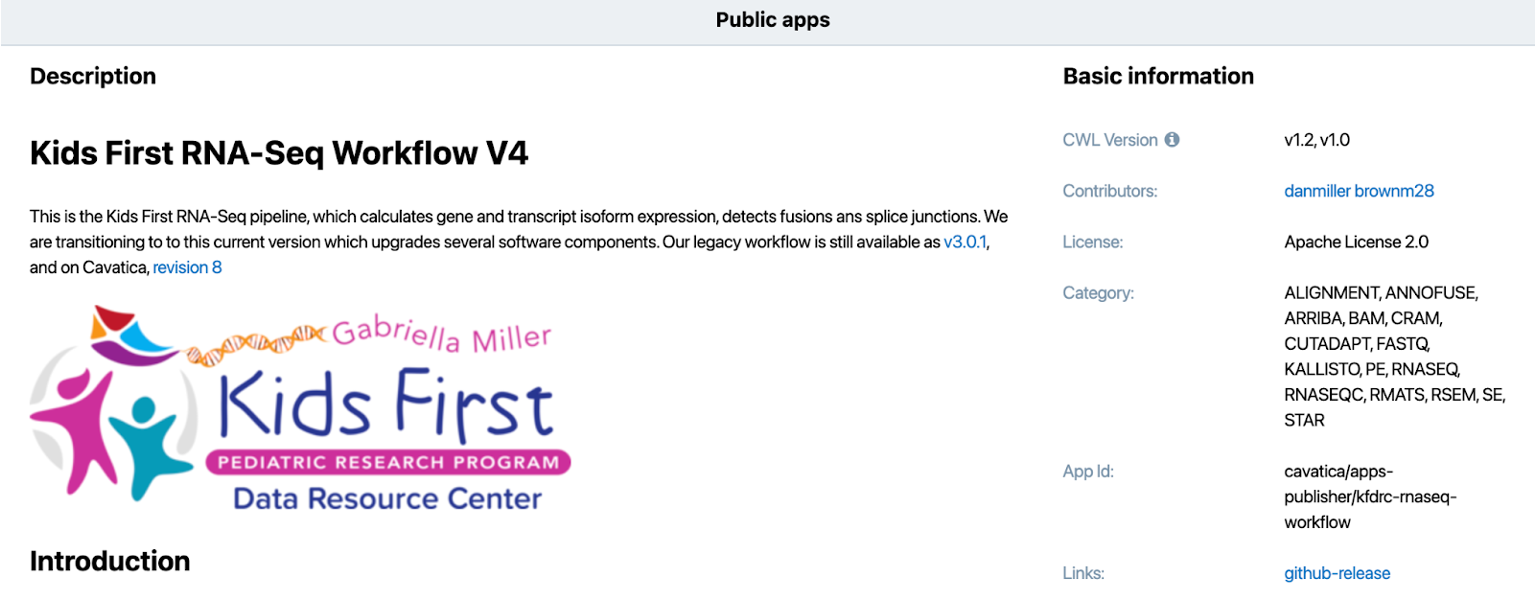
Importing the CFDE manifest via DRS into the CAVATICA workspace obviates the need to create a secondary copy of this data and improves security with authentication and authorization at time of access. The data from GTEx and Kids First were harmonized with the Kids First RNA-Seq Workflow public app.
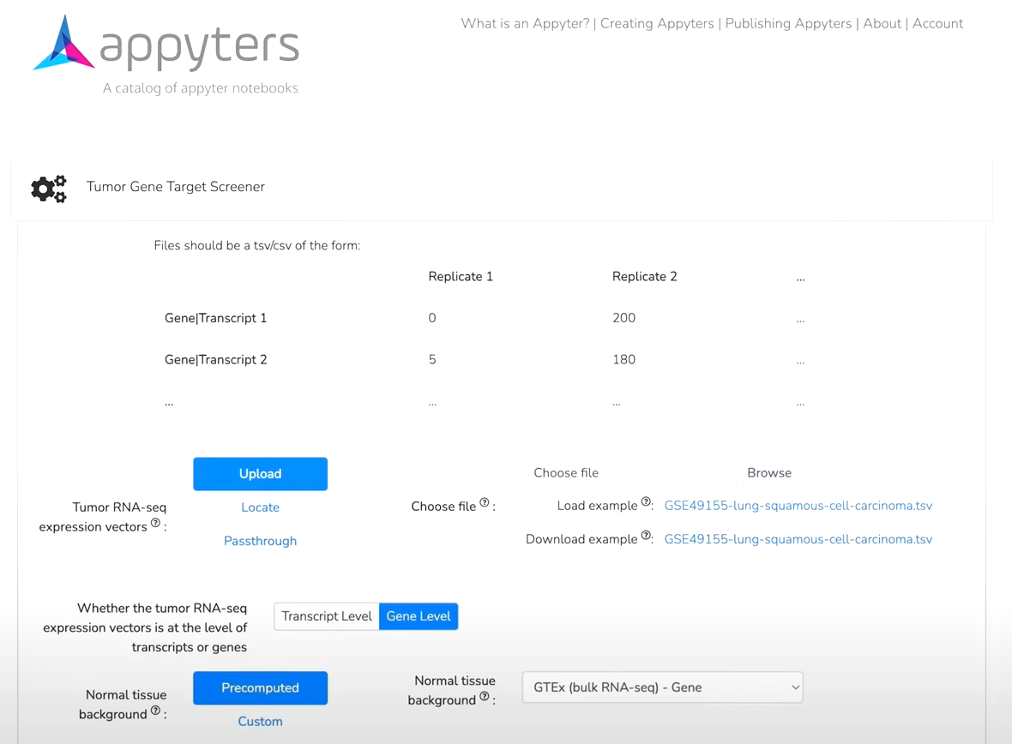
To showcase this capability, we use Tumor Gene Target Screener Appyter, specifically designed to do the exact same analysis as highlighted in a cancer cell paper reviewing neuroblastoma and GTEx. Appyters are graphical web browser-based software applications that enable users to execute bioinformatics workflows without coding. The goal is to confirm if this new analysis using the Appyters recapitulates the original findings of the cancer cell paper with this new larger cohort of neuroblastoma. Importantly, the Appyter can interoperate with and connect to the CAVATICA workspace where data were processed, harmonized, and structured. We find that many of the same genes that were initially identified were replicated in this analysis via the Appyter. Additional genes that were not initially in the list from the cancer research paper were also identified, including PHOX2A and PHOX2B, that were reported in a subsequent study published in Nature.
This work demonstrates how researchers can easily search for and seamlessly harmonize datasets across a federated DCC resource frameworks, without having to download data. Such federation and associated governance backed by GA4GH data exchange standards showcase a seamless integration across cohort analysis in a cloud-based environment. This accelerated democratized access can support comprehensive analysis and empower meaningful translational impact. Researchers can save time and reduce data fragmentation by utilizing this new DRS manifest-based solution.
Presentation
CAVATICA documentation: https://docs.cavatica.org/docs/import-from-a-drs-server
Slides: https://prezi.com/view/UBPyyMeP9cj1R3YUE6dT/
Video: https://www.youtube.com/watch?v=Z1hybV-V6ck
Poster: https://figshare.com/articles/poster/Manifest-based_DRS_import_A_practical_solution_for_cross-DCC_dataset_analysis_to_empower_translational_discovery_using_Kids_First_and_GTEx_data/21263148
Funding resources info:
NIH Common Fund Data Ecosystem (CFDE)
NIH Office of Data Science Strategy (ODSS)