Shifting towards personalized oncology
Powered by a wave of next generation sequence data from labs and the clinic, oncology is in the midst of a paradigm shift towards personalized medicine. To the modern cancer biologist, the genomic and transcriptomic features of a patient’s tumor have the potential to function as key guideposts for selecting an effective treatment regimen.
Clinical trials remain costly, however, and the need for reliable biomarkers for cancer drug sensitivity is as great as ever.
Exploring the Cancer Cell Line Encyclopedia
Enter the Cancer Cell Line Encyclopedia (CCLE), a collection of whole genome, whole exome, and RNA-seq datasets encompassing nearly 1000 human cancer cell lines. A project of the Broad Institute, Novartis Institutes for Biomedical Research, and the Genomics Institute of the Novartis Research Foundation, the CCLE correlates genomic data from 947 human cancer cell lines with pharmacological profiles of 24 anticancer drugs, allowing for large-scale comparative analysis.
| Experimental Strategy | Number of Samples | Number of Files |
| RNA-Seq | 935 | 1870 |
| Whole Exome Seq | 327 | 654 |
| Whole Genome Seq | 23 | 46 |
CCLE researchers selected a wide range of commercially available cancer lines from Europe, Japan, Korea, and the USA representing both much of the diversity of human cancer subtypes.
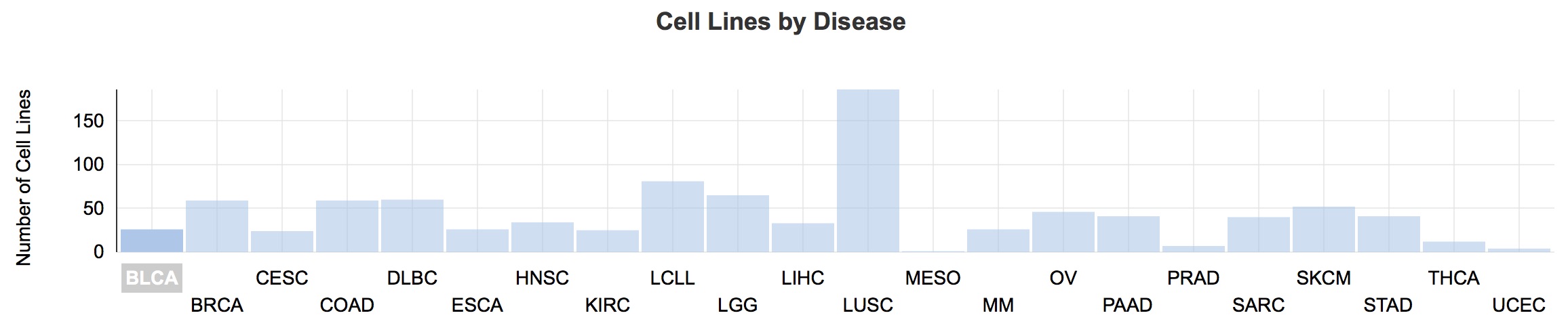
The number of cases for each cancer type (denoted by 2–4-letter codes) present in CCLE, viewed using the Cancer Genomics Cloud.
Each cell line was genetically characterized through a series of high-throughput analyses at the Broad Institute. Whole genome sequence data allows researchers to explore chromosomal rearrangements, exogenous sequence insertions, and mutations in lncRNAs and other intergenic elements, while high quality RNA-seq data allows researchers to dig deep into fusion genes and regulatory changes to the transcriptome.
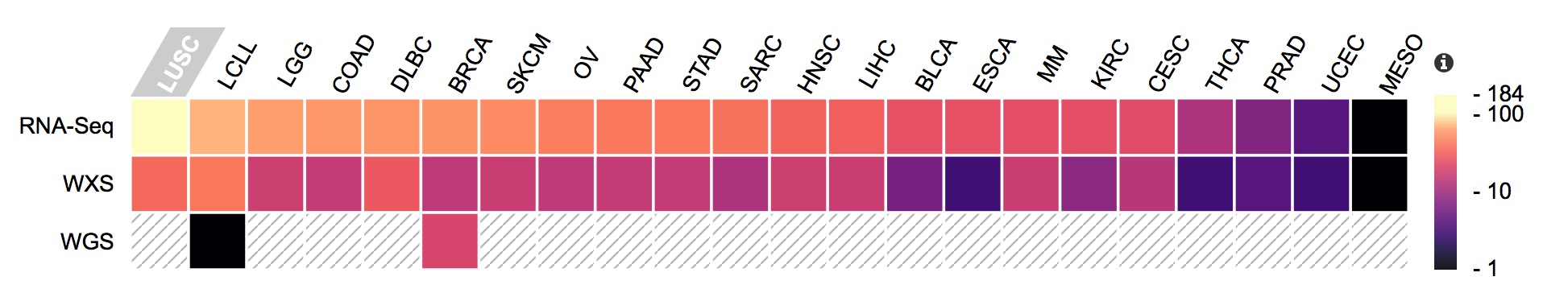
Molecular data present in the CCLE, with the number of available files represented as a heatmap.
Identifying molecular correlates of cancer drug response
In the five years since its initial publication, the CCLE has been validated against other large cancer datasets and used to identify numerous novel correlates of drug sensitivity. Indeed, the CCLE was critical in research correlating the efficacy of MEK inhibitors and topoisomerase inhibitors with specific transcriptional profiles in NRAS mutant cancers, identifying HLA expression as a biomarker for lymphocyte influx in triple negative luminal breast cancers, and developing the use of α-amanitin-antibody conjugates in the treatment of POL2A-negative cancers.
The CCLE is a valuable resource for anyone seeking to identify genetic correlates of drug response. It represents a systematic framework for investigating molecular correlates of drug response, allowing researchers to identify genomic features of clinical importance and use this information to accelerate clinical trial design.
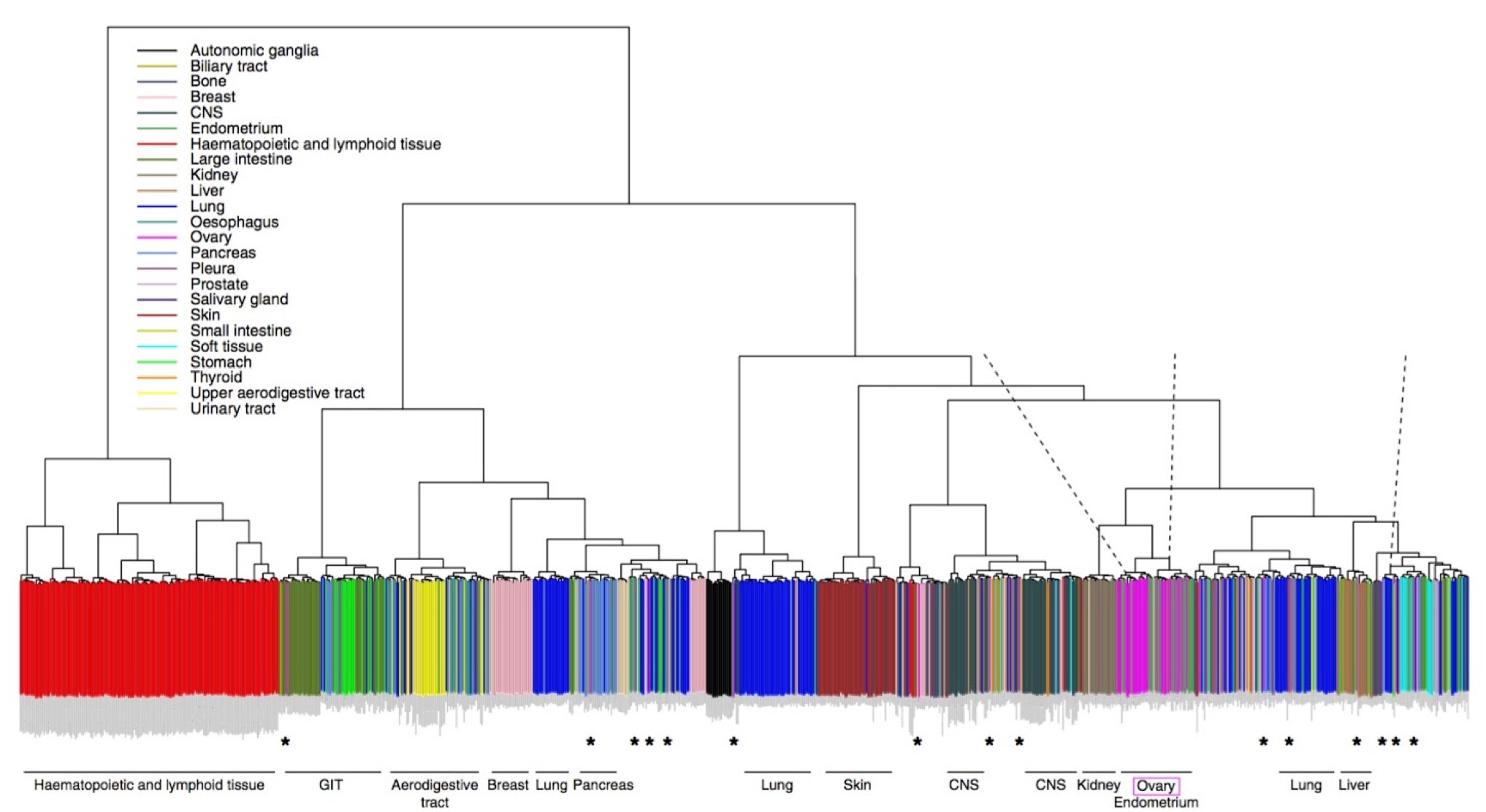
Expression-based clustering of all 963 CCLE cell lines. Adapted from Domke et al. Nature Communications 2013.
Accessing the data
Researchers can register at www.cancergenomicscloud.org to instantly explore and work with CCLE data alongside hundreds of bioinformatic tools, and on-demand access to thousands of CPU cores for analysis.