

Executive summary
Seven Bridges scientists and software supported the PanCancer Analysis of Whole Genomes (PCAWG) consortium when they urgently needed data analysis at large scale. Faced with over 800 TB of files to process, and with academic data centers unable to meet project deadlines, PCAWG engaged Seven Bridges to rapidly analyze their data. By deploying reproducible workflows in the cloud, our team quickly delivered variant calls for more than 1,350 cancer whole genomes.
- Seven Bridges bioinformaticians implemented BAM cleaning and variant calling workflows specified by PCAWG, using the Common Workflow Language to ensure reproducible analysis.
- We ran the analysis on the cloud-based Seven Bridges Platform, which automatically allocates compute resources to optimize analysis speed and cost effectiveness.
- We delivered output files that matched PCAWG requirements ahead of schedule, resulting in the allocation of additional samples for analysis.
PCAWG is a multinational research consortium requiring large-scale bioinformatics
The PanCancer Analysis of Whole Genomes (PCAWG) is an international collaboration from the International Cancer Genome Consortium (ICGC) investigating patterns of mutation in more than 2,800 patients with cancer. In late 2015, when conventional academic data centers were struggling to meet project deadlines, Seven Bridges joined the PCAWG technical working group, bringing our extensive experience in reproducible cloud-based bioinformatics and cancer genomics to bear on the data.
PCAWG includes patient donors from 48 cancer research projects with 20 different primary tumor sites (Figure 1). The project is designed to explore the consequences of somatic and germline variations in both coding and noncoding regions, with specific emphasis on cis-regulatory sites, noncoding RNAs and large-scale structural alterations (Box 1). PCAWG builds on previous work examining cancer coding regions across 12 tumor types from The Cancer Genome Atlas to develop an integrated picture of commonalities, differences and emergent themes across tumor lineages.1
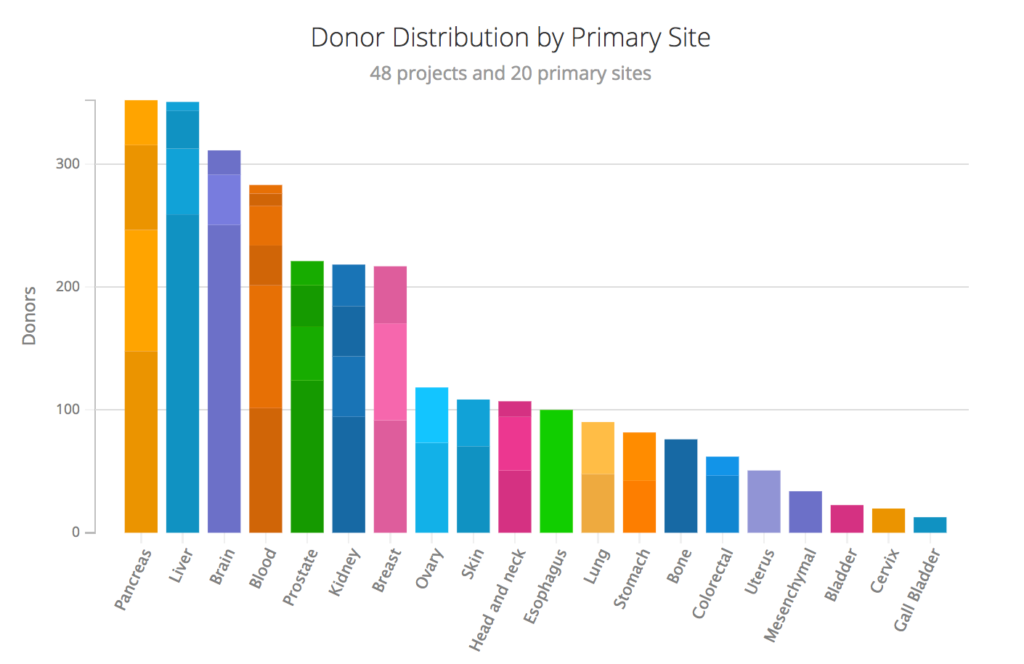
Figure 1 | Donor distribution by primary site. PCAWG includes donors from 48 different cancer research projects with 20 different primary tumor sites. Image from https://dcc.icgc.org/pcawg
For PCAWG, tumor and matched normal samples from each donor have undergone whole-genome sequencing (totalling 8,721 BAM files and requiring 800.9 TB of storage). A uniform set of alignment and variant calling algorithms was applied to each sequenced genome to ensure consistency of analysis among samples from the disparate research projects.
Box 1 | PCAWG Key areas of study
- Discovering driver mutations outside of the protein-coding regions of the genome;
- Integrating mutational signatures across tumor types and mutation categories;
- Characterizing subclonal structures and patterns of genome evolution across cancers;
- Investigating relationships between germline and somatic mutations;
- Investigating biological pathways targeted by driver mutations.
PCAWG enlisted Seven Bridges to accelerate their analysis
Realizing that academic data centers were unable to meet project deadlines, PCAWG sought industry partners with expertise in cloud-based computation and large-scale cancer genomics. Members of the consortium calculated that cloud services offered a faster and cheaper way to store and analyze their data.2 The consortium identified Seven Bridges as an ideal analysis partner, and the company joined the PCAWG technical working group in October 2015.
“[PCAWG] demonstrates how much faster and cheaper it is to use cloud computing than to use conventional academic data centers when analyzing vast biological data sets.” Stein, L. et al. (2015)
After joining the project, our team of scientists and bioinformaticians set to work to understand and implement the analysis workflow, in order to deliver the required output files as quickly as possible.
Seven Bridges bioinformaticians implemented complex reproducible workflows
To accelerate the PCAWG analysis, Seven Bridges scientists implemented and ran a PCAWG variant calling workflow rapidly and at large scale, to provide the required outputs within a matter of weeks. This workflow returned multiple VCF outputs for different variant callers, along with packaged intermediate files and associated metadata.
Once the consortium shared their aligned BAM files, our bioinformaticians implemented specified BAM cleaning and variant calling workflows. As with all analyses on Seven Bridges systems, these workflows are completely reproducible, ensuring uniform analysis across the samples, including those samples processed elsewhere at academic data centers. Along the way, our team identified and fixed a problem whereby one tool in the specified workflow occasionally and unpredictably used too much memory, which caused the whole workflow to fail silently.
Our team rapidly analyzed thousands of cancer whole genomes
We ran the analysis on the Seven Bridges Platform, the cloud-based system for biomedical data analysis. The Platform allocates storage and compute resources on demand to meet the needs of a given analysis. While the Platform runs on multiple infrastructures, for PCAWG we leveraged the ICGC data set hosted on Amazon Web Services (AWS), which allowed us to quickly, securely and cost-effectively access ICGC data hosted on AWS. The PCAWG analysis made extensive use of the Platform’s Application Programming Interface (API), which lets users build automated workflows to perform quality control checks, run analysis tools, and simplify the upload and metadata capture process.
Our team described these analytic tools using the Common Workflow Language (CWL) specification to ensure a reproducible, portable and scalable workflow. CWL, which Seven Bridges helps to develop, is a set of open-source specifications that also records version numbers and parameter settings. CWL tools and workflows are fully portable and not locked into any execution environment. For a project such as PCAWG in which standardization of output matters, this level of reproducibility is essential.
We initiated the analysis as soon as we obtained access to the relevant files and permissions. After validating the workflow on a subset of 20 samples from 4 donors, our team was able to rapidly scale up production to run batches of 300–600 tasks simultaneously on the cloud. By using the Seven Bridges Platform, we could automatically schedule tasks and allocate computation resources to ensure optimum speed and cost effectiveness.
The PCAWG samples were allocated across AWS machine types according to file size, which allowed optimization for cost efficiency. Because instances are priced differently depending on the amount of disk space and memory, this resulted in cost savings for the user.3
We delivered verified files matching the specified metadata scheme
The resulting output files were labeled according to a specified scheme and uploaded to a data center at the University of Chicago for inclusion in the final PCAWG data set (Figure 2).4 For each donor, we verified and uploaded more than 40 files comprising the VCF outputs from the specified workflows, packaged intermediate files and associated metadata.
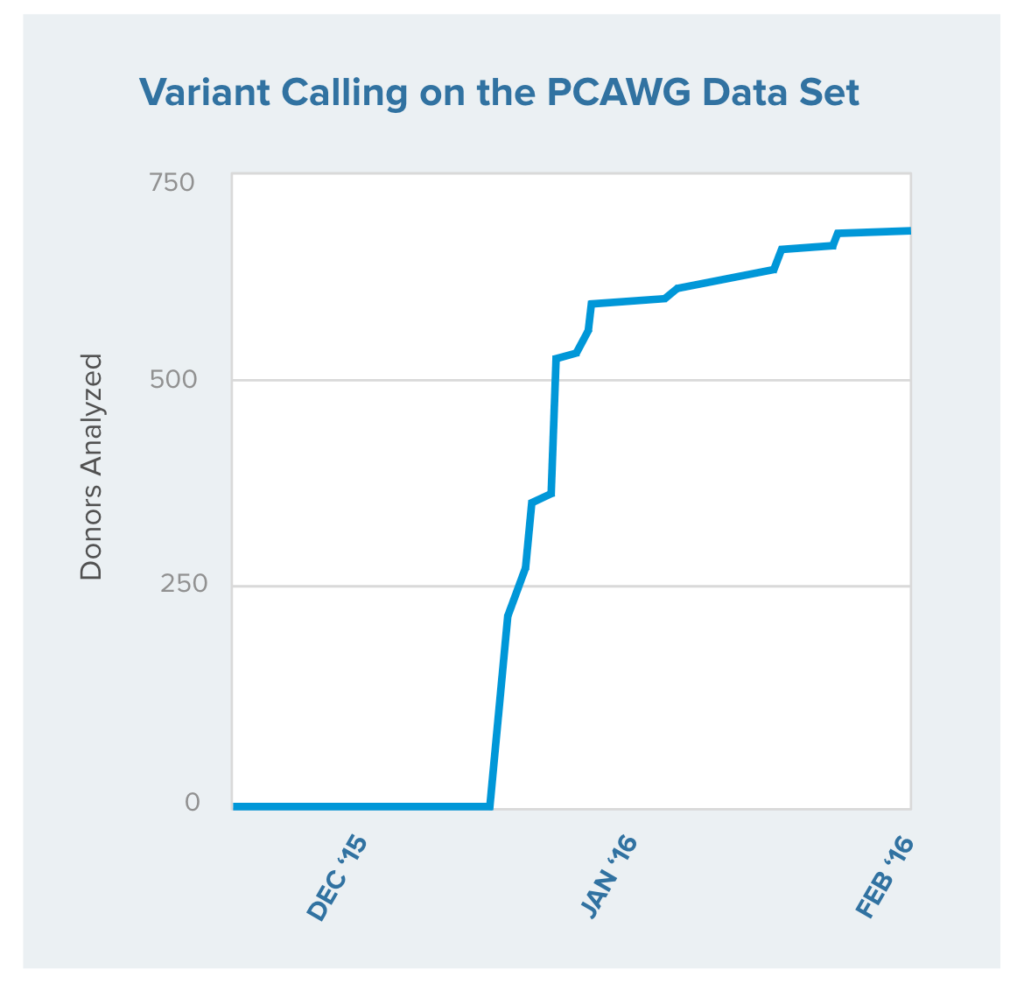
Figure 2 | Variant calling for the PCAWG data. The plot shows the rate at which the files from each of 655 donors were uploaded and verified. In total we delivered multiple variant calls from more than 1,350 cancer whole genomes in a matter of weeks.
The PCAWG consortium provided detailed instructions for the post-processing of output files, packaging into directory structure and syncing to the University of Chicago servers. To make file delivery as efficient as possible, our team of bioinformaticians worked with the PCAWG consortium to define dedicated output scripts to fit the specific metadata schema and requirements of production-grade processed data that will be used in downstream analyses by researchers worldwide.
By rapidly processing the samples in the cloud we returned the files ahead of schedule, with the result that the consortium allocated us additional samples for analysis. Thanks to the reproducible workflows described in CWL we were able to apply exactly the same analytic workflows to the additional allocation. In total Seven Bridges delivered variant calls for more than 1,350 cancer whole genomes to the final PCAWG project data set.
Our outputs support the next generation of cancer genomics research
By partnering with Seven Bridges, the PCAWG consortium was able to meet its aggressive project deadlines. Our expertise in reproducible bioinformatics meant we were able to provide official files for downstream analysis, which researchers worldwide will use to better understand cancer at the molecular level.
Following successful implementation and validation of the workflow, we completed the analysis of our allocated genomes in a matter of weeks, before applying the same workflows to an additional allocation. Overall, Seven Bridges delivered variant calls for more than 1,350 cancer whole genomes to the final PCAWG project data set where they will contribute to the next generation of cancer research.
1 The Cancer Genome Atlas Research Network et al. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45, 1113–1120; http://www.nature.com/ng/journal/v45/n10/full/ng.2764.html
2 Stein, LD et al. (2015) Data analysis: Create a cloud commons. Nature 523, 149–151; doi:10.1038/523149a
3 Estimated >20% savings on computation cost, based on Seven Bridges analysis.
4 Data available through IGCG Data Portal; https://dcc.icgc.org/pcawg