Welcome to the Cancer Genomics Cloud
Early Adopters
Welcome to the Seven Bridges Cancer Genomics Cloud.
Early access for researchers starts today.
The CGC helps scientists by hosting one of the world’s largest genomic datasets — The Cancer Genome Atlas (TCGA) — together with the analytical tools and computational resources needed to analyze it, on a secure, cloud-based platform.
This is important because cancer genomics is increasingly data-intensive. We can readily generate the genetic code of an individual (and their tumor) but extracting actionable insights from raw sequencing data can take days or months, even when split across hundreds of computers.
The CGC (www.cancergenomicscloud.org) is a complete cancer genomics data analysis ecosystem that Seven Bridges built as a part of the National Cancer Institute’s Cancer Genomics Cloud pilot program. [1] The overarching goal of the project is to develop and explore innovative methods for accessing and computing on large-scale genomic data. Making this data available in the cloud presents a unique set of challenges because genomic information is inherently personally identifiable. As a NIH Trusted Partner, Seven Bridges can authenticate and authorize approved researchers for TCGA data access in the cloud.
These technicalities aside, our mission for the CGC is tremendously simple: researchers with a hypothesis on cancer genetics can get started on their work immediately. From any computer. Anywhere. No campus cluster, or core facility, or waiting required.
Navigating one of the world’s largest genomics datasets
Initiated in 2005, TCGA is an immense and diverse gift to the global cancer research community; and more importantly, the patients who will benefit from what the data helps us learn about the disease. It’s this data that is at the core of today’s version of the CGC.
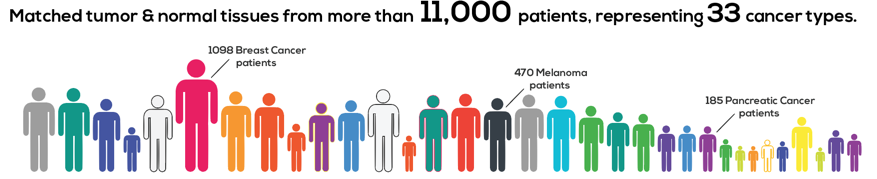
Thousands of cancer patients have donated tissue for analysis, and researchers around the United States examined it across multiple molecular dimensions (genomic, epigenomic, transcriptomic and proteomic analyses). This work generated more than a petabyte of raw data across hundreds of thousands of files. And in cancer research, it is clear that our understanding of an individual’s tumor is improved when we put it in the context of millions of others.
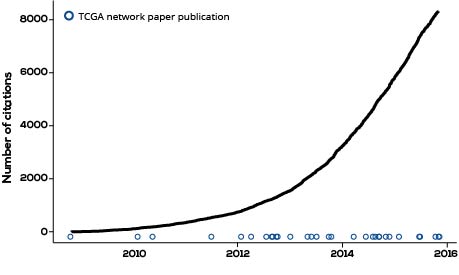
Though TCGA is only the beginning of this challenge, it has already created prodigious knowledge. Computational analyses of these data have led to substantial advances in our understanding of the molecular underpinnings of cancer. Hundreds of researchers participating in The Cancer Genome Atlas Network have published more than 20 ‘marker’ papers since the project began. These marker papers are, themselves, hugely important documents: they are academic lighthouses that deeply characterize an individual type of cancer and provide a roadmap for future researchers.
However, because one of the core principles of TCGA is to share data, even prior to publication,2 these marker papers represent a tiny fraction of the overall learning. To date, more than 8,000 papers have cited at least one of these marker publications. And the number of citing papers is accelerating over time.
Data alone is not enough
One guiding principle that we’ve followed in building the CGC is that making data available doesn’t make it useable. You can read more about how we’ve made TCGA data more findable, accessible, interoperable and reusable in the extensive CGC documentation. What we’re most proud of is what researchers can do once they’ve found the data they want: they can immediately start with predefined analytical workflows, modify or build a new one, add their own data, and then actually run their computation. Until now doing this work wasn’t anywhere near instant, and was only available to researchers in labs had access to computation and storage infrastructure that costs millions of dollars a year to own and operate.
Even better, we’ve worked with Amazon Web Services to make the dataset available through their Public Data Set program which fully covers the cost of storing TCGA data. This means that we’re able to use more of our Federal project funding to provide the research community with computation credits so that you can analyze and learn from these data for free.
What’s next: The Full Cancer Genomics Cloud in January
Although we’re releasing the CGC to approved users (apply here!) today, this is just the beginning. We believe that the only way to build software for the biomedical community is to both be a part of, and build with it. This is why we are opening our CGC early, and it’s why we’re hosting weekly office hours to answer questions and then improve the system. And, there’s a nine-month evaluation period, starting in January. In that time, we want everyone (including us!) to both kick the CGC’s tires, and use it for real research. We know that we will discover new use cases, and ways to make existing ones even easier.
It’s been 14 months since the Seven Bridges team started work on actually bringing the CGC to life, and being the one to write on behalf of the hundreds of people who helped it come to be is a huge honor. In addition to our teams in bioinformatics, design, engineering, quality control, and security, a great group of people at other organizations have been hard at work, too. Thanks to our friends at the National Cancer Institute, Amazon Web Services, The Broad Institute, the Institute for Systems Biology, and the Genomic Data Commons.
[1] This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN261201400008C.
[2] Consistent with the Fort Lauderdale data sharing principles