Cancer Cell Line Encyclopedia — Five Key Discoveries
Towards precision medicine
Powered by a wave of next generation sequence data from labs and the clinic, oncology is in the midst of a paradigm shift towards personalized medicine. To the modern cancer biologist, the genomic and transcriptomic features of a patient’s tumor have the potential to function as key guideposts for selecting an effective treatment regimen.
Clinical trials remain costly, however, and the need for reliable biomarkers for cancer drug sensitivity is as great as ever.
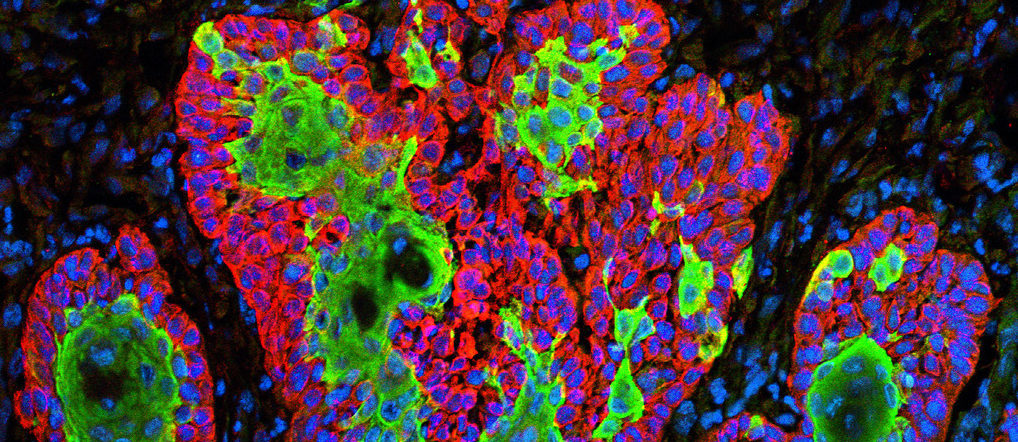 Micrograph showing the uncontrolled growth of cells in squamous cell carcinoma, the second most common form of skin cancer. Image by Markus Schober and Elaine Fuchs, The Rockefeller University
Micrograph showing the uncontrolled growth of cells in squamous cell carcinoma, the second most common form of skin cancer. Image by Markus Schober and Elaine Fuchs, The Rockefeller University
The idea of a large scale drug treatment dataset was brought to fruition in 2012 with the publication of the Cancer Cell Line Encyclopedia, which collected data on nearly 1000 human tumor cell lines, cancer cells which have been coaxed to grow indefinitely in the lab. A project of the Broad Institute, Novartis Institutes for Biomedical Research, and the Genomics Institute of the Novartis Research Foundation, the CCLE correlates genomic data from 947 human cancer cell lines with pharmacological profiles of 24 anticancer drugs, allowing for large-scale comparative analysis. Designed to represent much of the diversity of human cancers, the CCLE includes data from both common and rare cancer types.
Each cell line was genetically characterized through a series of high-throughput analyses at the Broad Institute, including whole genome, whole exome, and RNA sequencing.
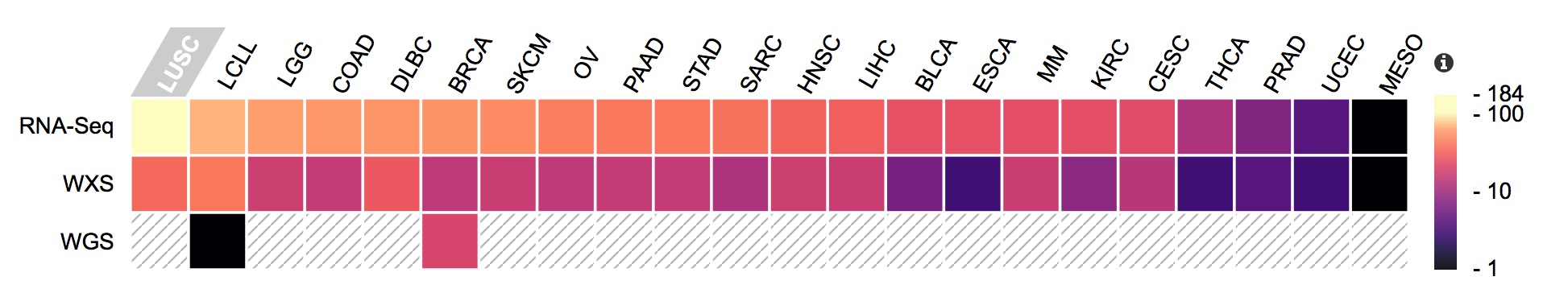 Molecular data present in the CCLE, with the number of available files represented as a heatmap.
Molecular data present in the CCLE, with the number of available files represented as a heatmap.
In the five years since its initial publication, the CCLE has been validated against other large cancer datasets and used to identify numerous novel correlates of drug sensitivity.
CCLE in the literature
Let’s take a look at how researchers have used the CCLE to expand the repertoire of oncogenic mutations and identify new routes for personalized cancer medicine.

Barretina et al. Nature 483, pp. 603–607 (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
This is the original paper that launched the CCLE. In addition to describing the dataset in depth, the authors give a nice proof of principle in identifying specific transcriptional profiles in NRAS mutant cancers that correlate with sensitivity to MEK inhibitors and topoisomerase inhibitors. This points to the value of the RNA-seq component of the dataset – although sequence variation at the DNA level is often in the limelight, gene expression can provide crucial information on the inner workings of cancer cells.
Huang et al. Science Vol. 339, Issue 6122, pp. 957-959 (2013). Highly recurrent TERT promoter mutations in human melanoma. Also see Huang et al. Abstract in AACR 103rd annual meeting proceedings. Single agent activity of PIK3CA inhibitor BYL719 in a broad cancer cell line panel.
Cancer cells are hotbeds for mutation. In addition to mutations within genes that can change the structure of important proteins, mutations can affect regulatory regions of the genome as well. In this Science paper, Huang et al. show that many cancers in the CCLE have specific mutations in the upstream regulatory region of the TERT gene, which codes for telomerase reverse transcriptase, an enzyme which is crucial for maintaining chromosome ends and is often implicated in aging. The authors showed that the mutations they identify can create a new binding site for specific classes of transcription factors, which bind to the mutated TERT gene and increase its expression two- to four-fold. Increased telomerase activity in these cancers may contribute to their immortal phenotype.
Huang’s group also used the CCLE to identify several molecular markers of for sensitivity to the cancer drug NVP-BYL719, including mutation of PIK3CA and copy number gains in ERBB2 and PIK3CA. Additionally, the authors identify mutations in the PTEN and BRAF as molecular signatures of NVP-BYL719-resistant cancers.
Domcke et al. Nature Communications 4, (2013). Evaluating cell lines as tumour models by comparison of genomic profiles.
Lack of a physiological microenvironment means that cultured cells are not a perfect stand-in for real tumors. This study addresses the standing question of how well cell lines growing in laboratory conditions represent those in tumors, using copy number and RNA-seq data from ovarian cancer lines in the CCLE. Their results show that some lines are better than others. Indeed, some of the most commonly used lines have become hypermutated compared to the cells in many ovarian tumors. The authors recommend an approach to selecting cell lines for in vitro studies that is informed by a better understanding of their underlying genetics.
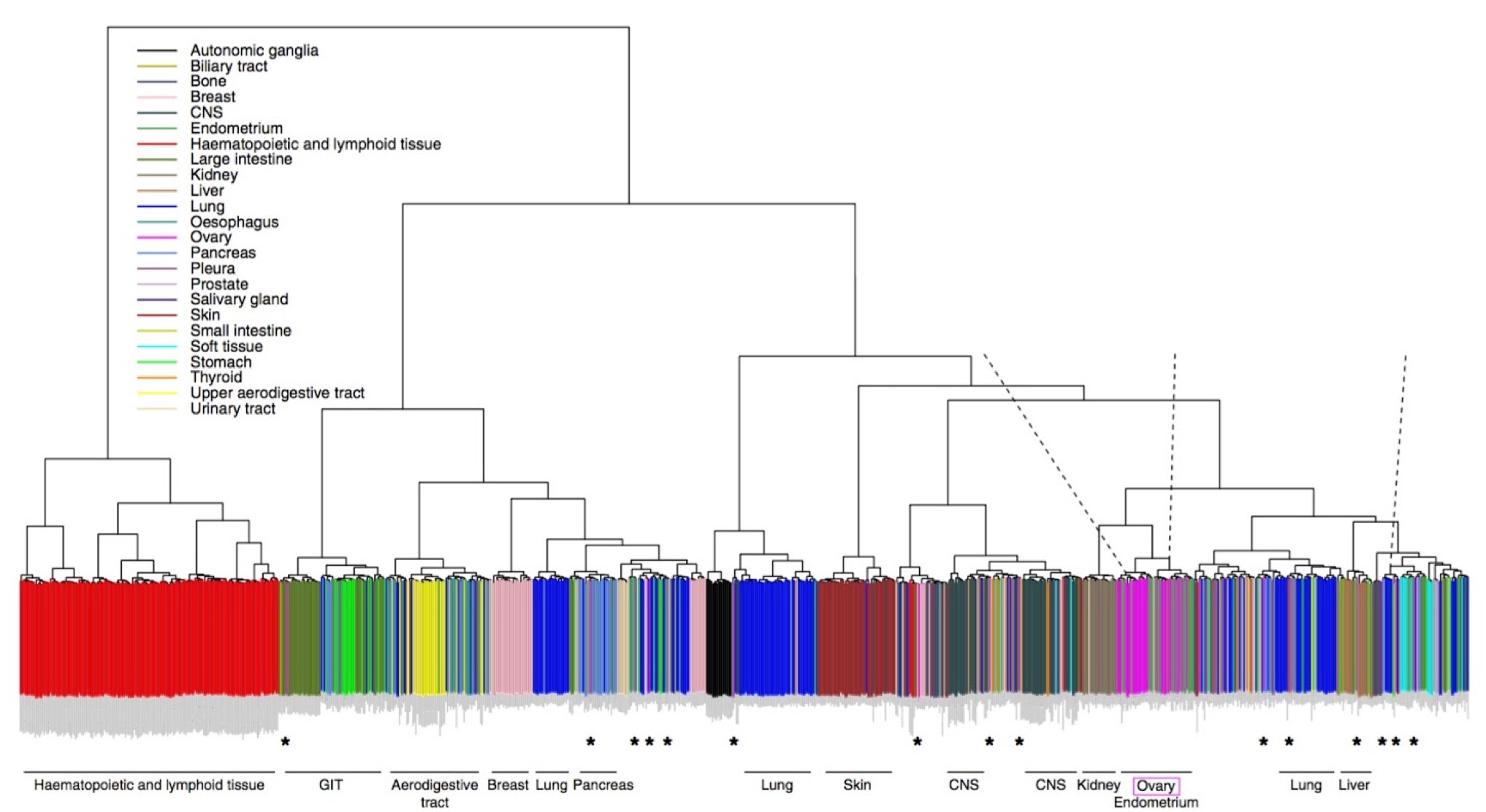
Expression-based clustering of all 963 CCLE cell lines. Adapted from Domke et al. Nature Communications 4, (2013).
Liu et al. Nature 520, pp. 697–701 (2015). TP53 loss creates therapeutic vulnerability in colorectal cancer.
Mutation of the tumor-suppressor gene TP53 is a common feature of cancer. Deletion of the TP53 locus can affect nearby genes as well. This study uses CCLE uncover the extent of deletions of the nearby POL2A gene in TP53-negative cancers and explores the use of α-amanitin-antibody conjugates in the treatment of these cancers.
Wilson et al. Gynecologic Oncology 143, Issue 1, pp. 143–151 (2016). Panobinostat sensitizes cyclin E high, homologous recombination-proficient ovarian cancer to olaparib.
This study uses CCLE data to develop preclinical evidence for the efficacy of the histone deacetylase inhibitor panobinostat against homologous recombination-proficient and cyclin E-overexpressing ovarian cancers.
Accessing the data
The CCLE dataset in its entirety is ready for researchers to use on the Seven Bridges Platform and through the Cancer Genomics Cloud.