Bioinformatics applications in cancer immunotherapy
An emerging characteristic, or hallmark, of cancer cells is their ability to evade destruction by the immune system. While immune surveillance plays a role in controlling tumorigenesis and tumor progression, cancer cells can in turn disable the immune system by secreting immunosuppressive factors or recruiting immunosuppressive cells. Because of this interaction between the immune system and cancer, there is growing interest in the biopharmaceutical industry to develop cancer immunotherapies.
Cancer immunotherapies can be broadly categorized into four classes:
- Therapeutic antibodies target surface molecules on cancer cells, and can either act alone or be conjugated to a toxic moiety;
- Whole cell therapies involve removing immune cells from a patient, engineering them to specifically target cancer cells, then transferring them back to the patient;
- Immune system modulators are small molecules, such as interferons and interleukins, that provide an unspecific boost to the immune system;
- Cancer vaccines deliver antigens to patients in order to stimulate an immune response.
One commonality among these approaches to immunotherapy is that they all require powerful, reproducible bioinformatics. Below, we highlight two specific examples of bioinformatics applications from the recent cancer immunotherapy literature.
1. Selecting neoantigens for personalized vaccines
A contributing factor to tumor heterogeneity, de novo somatic mutations frequently arise in cancer cells. Because the mutations may be absent from the normal human genome, peptides derived from mutated regions of the cancer genome can be immunogenic if presented by major histocompatibility complex (MHC) molecules. These so-called neoantigens can be administered as a vaccine in order to elicit a T cell response against the tumor.
A 2014 preclinical study demonstrated the power of using bioinformatics to identify neoantigens. First, whole exome and RNA sequencing were performed on mouse tumor cells. By comparing the whole exome data to the mouse reference genome, the researchers identified mutations specific to the cancer cells. They then used RNA sequencing to confirm which mutations were actually expressed by the cells.
To evaluate the immunogenicity of the mutated peptides, the researchers immunoprecipitated MHC molecules and analyzed them with mass spectrometry to determine which peptides were presented as epitopes. An algorithm was then used to predict interactions between the putative epitopes and T cell receptors. The final subset of mutated peptides was administered to mice, and the resulting CD8 T cell response against tumor growth showed that the peptides were productive neoantigens.
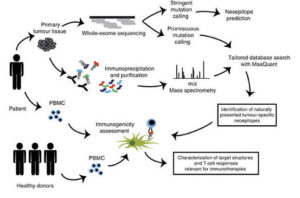 A pipeline to predict neoantigens from whole-exome sequencing data and evaluate their immunogenicity. Adapted from Bassani-Sternberg et al. 2016.
A pipeline to predict neoantigens from whole-exome sequencing data and evaluate their immunogenicity. Adapted from Bassani-Sternberg et al. 2016.
As neoantigen vaccines undergo further development for cancer patients, bioinformatics pipelines will play a key role in the technology for one major reason: most tumor mutations are patient-specific. As a result, sequencing must be performed on the tumor and normal cells of each patient in order to identify their unique neoantigens. A robust pipeline to analyze sequencing data and call variants will serve as the foundation of these personalized vaccines.
2. Understanding the effectiveness of immune checkpoint inhibitors
The checkpoint pathways that regulate immune cells can be co-opted by cancer cells to evade immune attack. For example, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) is a checkpoint receptor that, when bound, downregulates the immune response. Because some cancer cells upregulate CTLA-4 expression, a successful immunotherapy strategy is to inhibit CTLA-4. One such therapy, the monoclonal antibody ipilimumab, has demonstrated long-term clinical benefit for 20% of treated patients with metastatic melanoma. In order to better understand the patient predictors of clinical benefit, a 2015 study employed a number of genomics approaches.
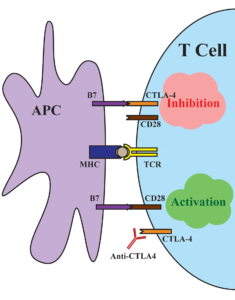 The mechanism of how CTLA-4 acts as an immune checkpoint. When bound to its normal ligand B7, T cell immune response is inhibited. When bound to an antibody such as ipilimumab, immune response is activated. Adapted from Su and Fisher 2016.
The mechanism of how CTLA-4 acts as an immune checkpoint. When bound to its normal ligand B7, T cell immune response is inhibited. When bound to an antibody such as ipilimumab, immune response is activated. Adapted from Su and Fisher 2016.
In the study, researchers examined a cohort of metastatic melanoma patients who received ipilimumab treatment. Each patient provided a pretreatment tumor biopsy sample as well as a control germline sample. Whole exome sequencing performed on the samples showed that clinical benefit was associated with increased numbers of mutations and neoantigens in the tumor cells, although no patterns were identified among the specific neoantigen sequences associated with clinical benefit. RNA sequencing showed that higher expression of cytolytic genes, such as granzyme A and perforin, was associated with clinical benefit.
Other studies have shown similar results for how tumor genomes impact the effectiveness of ipilimumab, and Boussiotis has proposed a mechanistic explanation for these observations. During the immune response, tumor-specific T cells are activated when neoantigens are presented. Activated T cells express more CTLA-4 on their cell surfaces, such that CTLA-4 is accessible to be bound by ipilimumab. If tumors do not have presented neoantigens, then tumor-specific T cells are not activated; CTLA-4 does not accumulate on T cell surfaces, so ipilimumab has no effect.
As the therapeutic field of immune checkpoint inhibitors matures, bioinformatics studies will be important to the drug development process. Bioinformatics analyses can provide insight into how current therapeutics work, as well as identify gaps to be addressed by future generations of therapeutics.
Impact of bioinformatics on cancer immunotherapy
Bioinformatics methodologies play an important role in all aspects of cancer immunotherapy development. In the case of neoantigen vaccines, bioinformatics is an essential part of personalizing the therapy to each patient. Bioinformatics can also be used to refine treatment, as is the case with studying why ipilimumab is only effective in a subset of patients.
As the field of cancer immunotherapy moves forward, implementing efficient and reproducible bioinformatics pipelines will be crucial to success. At Seven Bridges, we design our products—including the Seven Bridges Platform—based on these principles. Together, our products and team of experts are prepared to facilitate bioinformatics studies at all stages of drug development.